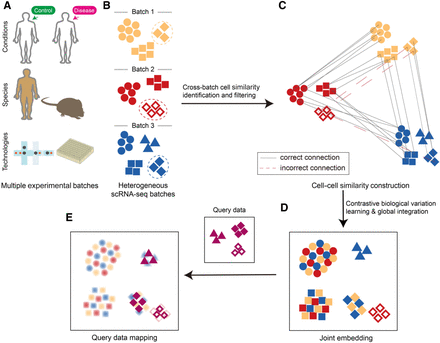
Overview of the scInt method. (A) scInt integrates multiple scRNA-seq batches that may come from different conditions, species, or technologies. (B) Three heterogeneous batches. Batches and cell types are represented by different colors and shapes, respectively. The dashed cell groups represent the same cell types with different transcriptomic states. (C) Cell–cell similarity construction. Similarities based on a cluster-specific exponential kernel are identified and further filtered in the cPCA space. (D,E) Integration and reference-based mapping of multiple batches. A unified contrastive biological variation learning framework is used for integration (D) and reference-based mapping (E).











