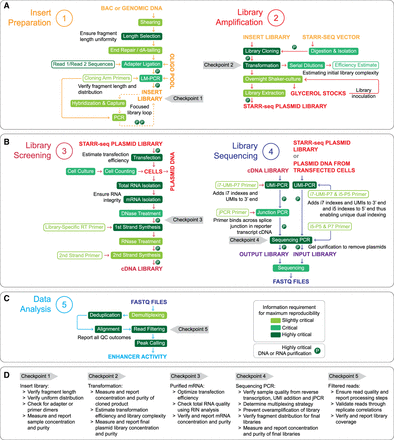
Roadmap to a successful STARR-seq assay. Schematic illustrates five major sections of STARR-seq: (A) Insert preparation and library amplification, (B) library screening and library sequencing, (C) data analysis. Individual steps in each section are categorized as slightly critical (light green), critical (green), or highly critical (dark green) based on the importance of reporting the methodological detail and intermediate results for a reproducible assay. (D) Assay checkpoints. Each section also has a checkpoint, which may serve as stopping points in the protocol to perform validation of previous steps before moving to the next step. Key QC measures that may be performed at these checkpoints are also provided in the bottom panel. Of note, panel B, section 4, “Library Sequencing,” illustrates a methodology for adding unique molecular identifiers (UMIs) and unique dual indexes. A more detailed schematic along with sequence information is provided in Supplemental Figure S1C–E, Supplemental Table S2, and Supplemental Protocol. Alternative methods for UMI addition have also been reported in several studies.











