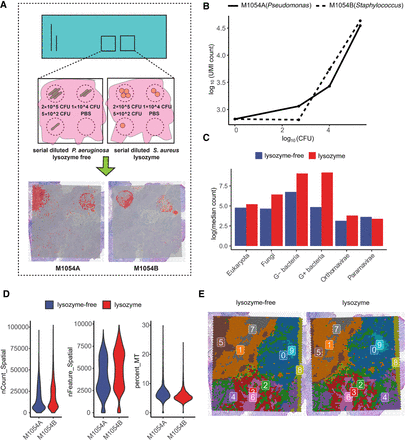
Capture efficiency and recall of SMT as measured by positive controls and evaluation of experimental optimization for SMT. (A) Study design of the positive control and optimization experiments and, shown at the bottom, spatial distribution of the microbial sequences captured by SMT superimposed on tissue histology, showing clear circular patterns. (B) Control bacteria sequences captured by SMT at each corner (measured as total microbial UMI count; y-axis) against actual count of bacteria applied at that corner (measured as CFU; x-axis). (C) Bar plot showing microbial sequence capture with or without the additional lysozyme step. (D) Violin plots of host gene UMI count, gene count, and mitochondria percentage (common measurements to QC single-cell and ST data) with or without lysozyme treatment, no significant difference could be observed. (E) Spatial dim plots showing clustering of spatial spots using host expression data with or without lysozyme treatment; the similar pattern between the two conditions indicated the effect introduced by lysozyme treatment on host gene expression was minimal.











