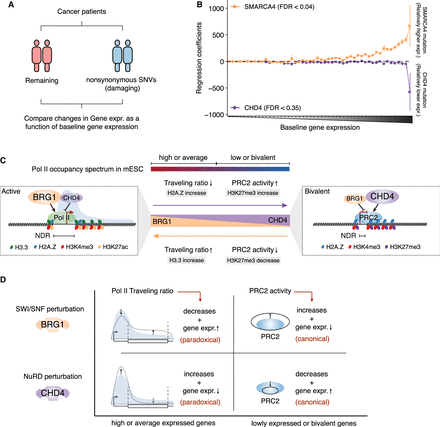
The impact of SMARCA4 and CHD4 lesions in cancer on gene expression. (A) Schematic illustration of the approach used to compare gene expression differences between patients having mutations in SMARCA4 or CHD4 to those of the remaining patients from 11 different cancer cohorts. (B) Mean regression coefficients for genes (N = 19,592 binned into 50 groups of 392 genes each and sorted by their expression levels) across 11 cancer cohorts along with standard error bars. Regression coefficients reflect a relatively higher (positive) or lower (negative) expression level of genes in patients having SMARCA4 or CHD4 mutations in comparison to those of the remaining patients. (C) A model explaining the antagonistic activities of SWI/SNF and NuRD chromatin remodelers at gene promoters during steady-state and MEF-to-mESC reprogramming. (D) Summary of the canonical and paradoxical impact of SWI/SNF and NuRD activities on gene expression following their perturbation.











