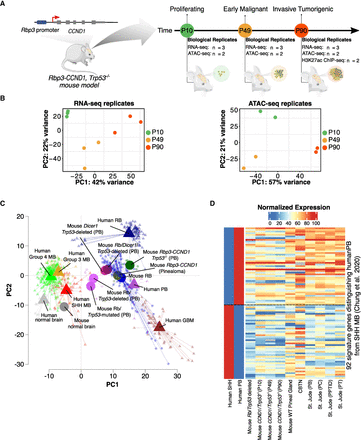
A mouse model recapitulating the evolving PB. (A) The Rbp3-CCND1/Trp53−/− mouse model and timeline representing the three key time points of PB progression, the assays performed, and the number of biological replicates. (B) PCA plots for RNA-seq and ATAC-seq samples used, highlighting the similarity between the biological replicates. (C) GSEA-PCA plot of different mouse model samples of PB and RB, as well as human normal brain, PB, MB, and GBM samples shows that the Rbp3-CCND1/Trp53−/− model is closer to other mouse models of PB, RB, and human PB but is further away from human RB and GBM. For each group, each sample or replicate is represented by a symbol of the same shape and color, albeit more transparent, as the corresponding centroid. Included also are mouse pineal samples with Rbp3-CCND1 that develop hyperplastic pineal glands but no malignant tumors (pinealoma). (D) Heatmap of normalized expression profiles of 92 signature genes distinguishing human PB from SHH MB across multiple human PB and other pineal tumors samples (Methods) as well as different mouse models of PB showing the closer similarity of Rbp3-CCND1/Trp53−/− to the Rb/Trp53-deleted mouse model as well as to human PB samples.











