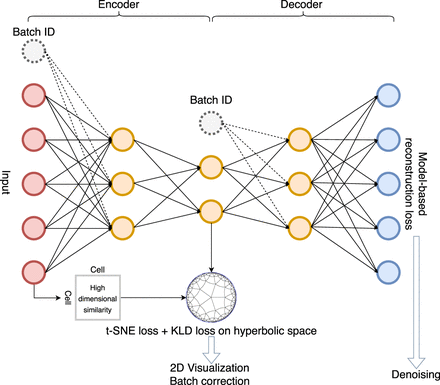
Network architecture of scDHMap. The encoder and decoder are fully connected neural networks. The latent embedding is in a 2D hyperbolic space for visualization and with the t-SNE regularization for structural preservations. KL divergence loss minimizes the divergence between the posterior and prior distributions of the latent embedding. Batch IDs can be incorporated to align different batches. Zero-inflated negative binomial (ZINB) reconstruction loss characterizes single-cell count data.











