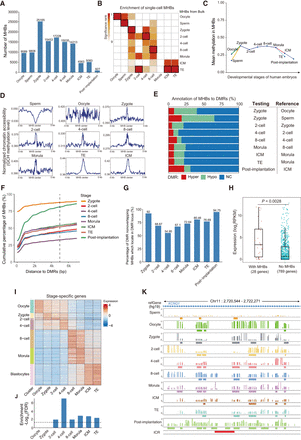
Regulatory importance of MHBs in human embryonic development. (A) Number of MHBs identified from bulk bisulfite sequencing of early human embryos. (B) Validation of MHBs using single-cell bisulfite sequencing data. An enrichment test was performed using the R package LOLA, with the union of single-cell MHBs as the test background. Each set of single-cell MHBs was tested against all sets of bulk MHBs; the cell type with the highest significance is highlighted by a black rectangle. (C) Temporal dynamics of mean methylation in MHB regions during early human embryonic development. (D) Normalized signals representing chromatin accessibility proximal to the center of MHB regions. (E) Annotation of MHBs to DMRs. DMRs were identified by comparing the test and reference groups. (Hyper) Hyper-DMR, (Hypo) hypo-DMR, (NC) no significant change. (F) Percentage of MHBs situated near DMRs. (G) MHBs tend to be located in DMR loci. Bar plots show the percentage of DMR-nonoverlapping MHBs that are located within DMR-defined loci. (H) The presence of MHBs in promoter regions is associated with gene activation. The expression of genes with and without MHBs was compared among genes with high methylation (>0.8) in promoters. Statistical significance was evaluated using the Wilcoxon rank-sum test. (I) A heatmap showing the expression of stage-specific genes. (J) Enrichment of MHBs in stage-specific genes in matched stages. The enrichment test was performed using the R package rGREAT. (K) An example genomic track displaying the known maternal imprinted control region KCNQ1. Mean DNA methylation and MHB tracks are shown for each stage in the region (Chr 11: 2,720,544–2,722,271).











