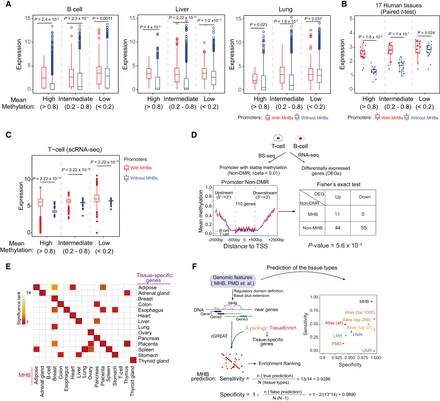
Tissue MHBs informative of gene expression. (A) Distribution of gene expression in promoters with different DNA methylation patterns in B cell, liver, and lung tissues. Promoters were categorized into low- (<0.2), intermediate- (0.2–0.8), and high-methylation (>0.8) groups. Expression was compared between promoters with and without MHBs within each group. Statistical significance was evaluated by the two-sided t-test. (B) The overall association of MHBs with gene expression across 17 tissue types was evaluated with the paired t-test. (C) MHB associations with scRNA-seq gene expression in T cells stratified by mean methylation levels. (D) MHBs are associated with differential gene expression. DMRs in T cells and B cells were identified using metilene (FDR < 5%, Δbeta > 0.1). The bottom left panel shows the average methylation around TSSs of differentially expressed genes with nearly identical DNA methylation in promoter regions (Δbeta < 0.01). The bottom right panel is a 2 × 2 contingency table categorizing genes by differential expression and MHB status in T cells. Statistical significance was evaluated using a Fisher's exact test. (E) Enrichment of MHBs with tissue-specific genes. Enrichment was tested using the R package rGREAT. The resulting adjusted P-values were ranked across all tissue types. Black rectangles indicate the top-ranked tissue with the highest significance. (F) Comparison of tissue type prediction performance for MHBs versus other methylation-associated genomic features. The left panel illustrates the prediction process using different genomic features. The right dot plot shows the prediction sensitivity and specificity.











