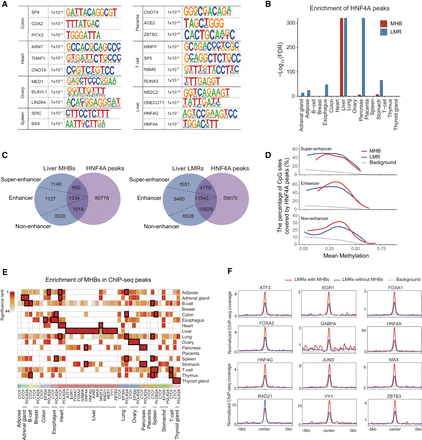
DNA motifs identified in tissue MHBs. (A) Representative DNA motifs enriched in tissue MHBs. HOMER was used to identify significantly enriched motifs in tissue-specific MHBs, with the union of MHBs as the test background. The top 10 motifs with P-values < 0.05 were retained for each tissue. (B) Enrichment of liver HNF4A ChIP-seq peaks in MHBs and LMRs. Enrichment tests were performed using the R package LOLA, with the union of MHBs and LMRs as the test background. To preclude computing the logarithm of zero, FDR values of zero were converted to 10−300 before logarithmic transformation. (C) Venn diagram showing the overlap between MHBs or LMRs and HNF4A ChIP-seq peaks in the liver. Liver MHBs and LMRs were categorized into three groups based on overlap with super-enhancers and enhancers (EnhA1). (D) Enrichment of HNF4A ChIP-seq peaks in liver MHBs is independent of the presence of known enhancers. Enrichment scores were defined as the percentage of CpG sites covered by open chromatin regions. Enrichment analysis was performed with the computeCpgCov function in mHapSuite. (E) Enrichment of MHBs with ENCODE ChIP-seq peaks. Enrichment tests were performed using the R package LOLA, with the union of MHBs as the test background. The resulting P-values for significant enrichment were ranked across all tissue types (FDR < 5%), and the top three are indicated by black rectangles. (F) Normalized transcription factor ChIP-seq signal around the centers of LMRs in liver tissue. ChIP-seq signals of transcription factors in liver LMRs with or without MHBs are shown in red and blue, respectively. To control for confounding factors, only regions with similar methylation levels (0.3–0.45) were considered.











