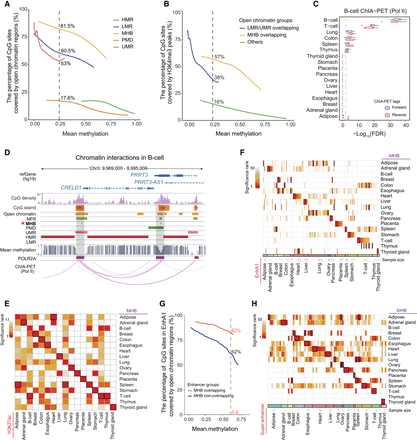
Tissue MHBs are enriched in regulatory elements. (A) Enrichment of open chromatin regions in MHBs as a function of mean methylation. The enrichment score was calculated as the percentage of CpG sites covered by open chromatin regions. (PMD) Partially methylated domain, (LMR) low-methylated region, (UMR) unmethylated region, and (HMR) high-methylated region. (B) Enrichment of H3K4me3 modification in open chromatin regions that overlap MHBs. Open chromatin regions were categorized based on overlap with UMRs/LMRs, MHBs, or neither. The enrichment score was calculated as the percentage of CpG sites covered by H3K4me3 ChIP-seq peaks. (C) Enrichment of B cell ChIA-PET (Pol II) tags in tissue MHBs. The enrichment was tested using the R package LOLA, with the union of MHBs from all tissues as the background. (D) An example genomic track showing that chromatin interactions tend to occur in MHB regions. Tracks of CpG density, mean methylation, MHBs, methylation-associated regions (PMD, LMR, UMR, and HMR), NFR, open chromatin regions, and chromatin interaction in this region (Chr 3: 9,969,000–9,995,000) for B cells are shown. (NFR) Nucleosome-free region. (E,F) Tissue-specific enrichment of MHBs in H3K27ac peaks and EnhA1 regions in matched tissue types. (G) Greater enrichment of open chromatin regions in enhancers with MHBs. The enhancers represented by EnhA1 regions were divided into two groups based on whether they overlap with MHBs. Compared with EnhA1 regions without MHBs, enhancers overlapping MHB regions are more enriched in open chromatin regions, regardless of the distribution of mean methylation. The enrichment score is calculated as the percentage of CpG sites covered by regions of open chromatin. (H) Tissue-specific enrichment of MHBs in super-enhancer regions in matched tissue types. Enrichment tests in E, F, and H were performed using the R package LOLA, with the union of MHBs from all tissues as the test background. The resulting adjusted P-values for significant enrichment (FDR < 5%) were ranked across all tissue types. The black rectangle highlights the top one tissue in E, the top five in F, and the top three in H. Nonsignificant (FDR > 5%) results are shown in white. Enrichment tests in A, B, and G were performed with the computeCpgCov function in mHapSuite.











