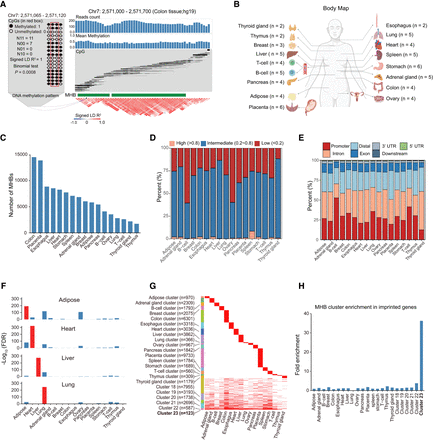
MHB landscapes identified in 17 representative human normal tissues. (A) Schematic of MHB patterns. The left panel shows an example of linkage disequilibrium (LD) between two CpG sites. A region (Chr 7: 2,571,065–2,571,120) with four CpG sites is depicted with a lollipop plot of 18 sequencing reads, in which black circles represent methylated cytosines and white circles represent unmethylated cytosines. The second and third CpG sites are comethylated, as indicated by the signed LD R2. Statistical significance was assessed using a binomial test. N11 indicates the number of MHAPs that are methylated at both CpG sites; the numbers of MHAPs of the other three types are also shown. The right panel shows an example of an MHB in colon tissue (Chr 7: 2,571,000−2,571,700). The top part shows the coverage and mean CpG methylation of each CpG site, and the middle part displays the DNA methylation status of the individual fragment, in which black and white represent methylated and unmethylated CpG sites, respectively. The bottom part shows the heatmap of the signed LD R2 score between pairs of covered CpG sites. (B) Body map showing analyzed normal tissue types and sample sizes. Panel created with BioRender (https://www.biorender.com). (C) Number of MHBs identified per tissue type. (D) Bar plots illustrating the proportion of MHBs in low (<0.2), intermediate (0.2–0.8), and high (>0.8) methylation groups. (E) Bar plots illustrating the proportions of MHBs annotated to different genomic features. (F) Validation of MHBs in four tissue types using independent data sets. Enrichment was determined by the R package LOLA, using the union of MHBs from all tissue types as the background. To preclude computing the logarithm of zero, FDR values of zero were converted to 10−300 before logarithmic transformation. (G) Categorization of MHBs into 23 nonoverlapping clusters. The number of regions in each cluster is shown. (H) A bar plot illustrating the enrichment of MHBs in the loci of imprinted genes, with fold enrichment evaluated using rGREAT.











