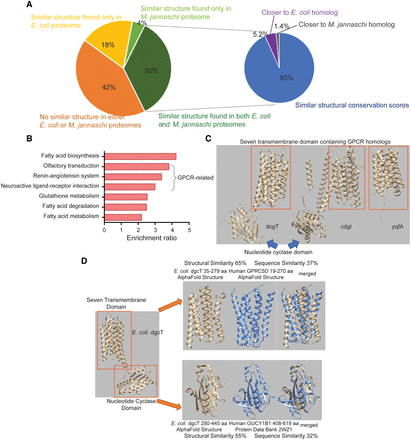
Global structural comparison sheds insight into the origin of the GPCR signaling pathway. (A) Distribution of structurally conserved and nonconserved proteins in the human proteome. Fractions of human proteins with structural homologs in the E. coli or M. jannaschii proteomes are shown. A sub-pie chart is presented for proteins with structural homologs found in both the E. coli and M. jannaschii proteomes. “Similar structural conservation scores” indicates a difference smaller than 0.1 between the scores, whereas “closer to E. coli homolog” or “closer to M. jannaschii homolog” indicates that the difference in the structural conservation score is greater than 0.1. (B) Gene overrepresentation analysis for proteins with a structural homolog found in E. coli only. For the list of proteins used for the overrepresentation analysis, see Supplemental Table S7. (C) The tertiary structures of the three GPCR-like proteins found in the E. coli proteome. The orange box indicates the seven-transmembrane domain. (D) Structural analysis for E. coli dgcT. The protein consists of two domains, one seven-transmembrane domain, and one nucleotide cyclase. The most structurally similar homolog for each domain is shown.











