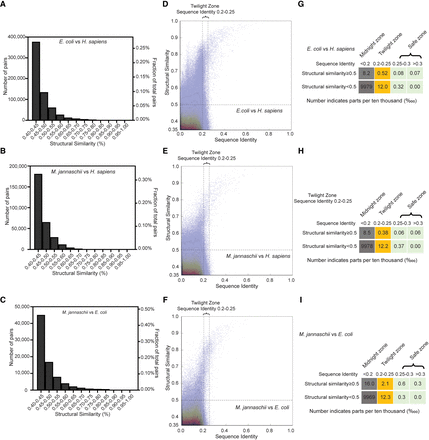
Distribution of structural similarity and sequence identity when comparing proteins at the proteomic scale. (A–C) Distribution of the numbers of protein pairs with structural similarity above 0.4 when comparing AlphaFold2 modeled proteins from one species with those from another. The subfigures compare the proteomes of E. coli versus H. sapiens (A), M. jannaschii versus H. sapiens (B), and M. jannaschii versus E. coli (C). Note that the number of protein-coding genes in each species differs, so a greater absolute number does not necessarily indicate a greater proportion. The left y-axis indicates the absolute number, and the right y-axis indicates the proportion in all possible protein combinations. (D–F) Overlay of 2D scatter plot and 2D contour plot depicting the distribution of structural similarity and sequence identity of the proteins. The subfigures compare the proteomes of E. coli versus H. sapiens (D), M. jannaschii versus H. sapiens (E), and M. jannaschii versus E. coli (F). The vertical dashed lines indicate the sequence identity of 0.2 and 0.25, and the horizontal dashed line indicates the structural similarity of 0.5. (G–I) The proportion of protein pairs of all possible combinations in each zone. The subfigures compare the proteomes of E. coli versus H. sapiens (G), M. jannaschii versus H. sapiens (H), and M. jannaschii versus E. coli (I). The twilight zone is defined as a sequence identity between 0.2 and 0.25. The safe zone is defined as sequence identity greater than 0.25, and the midnight zone is defined as sequence identity less than 0.2.











