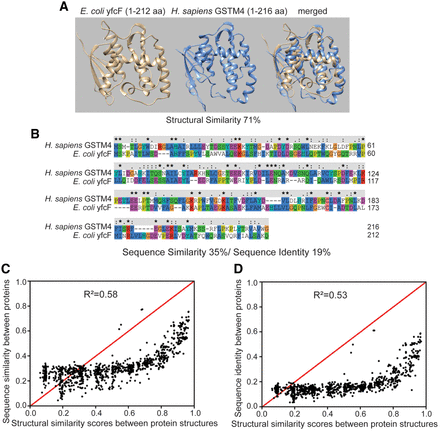
Comparison between structural similarity and sequence similarity or identity. (A,B) An example showing that proteins with nearly superimposable tertiary structures may have a low level of sequence similarity or identity. (A) AlphaFold2 modeled protein structures for E. coli and human glutathione S-transferase and alignments of the two structures. (B) Sequence alignment of E. coli and human glutathione S-transferase. The color indicates residue groups (blue, hydrophobic; red, positive charge; magenta, negative charge; green, polar). Sequence identity and similarity are indicated on top of the amino acid letters. An asterisk suggests a fully conserved position; a colon suggests strong similarity, and a period suggests weak similarity. (C,D) Comparison between structural similarity (TM-score) and sequence similarity (C) or identity (D) for 1000 pairs of selected experimental structures. The samples were obtained through stratified sampling, in which random sampling was performed at each stratum level (structural similarity ranging from x to x + 0.1, where x counts from 0.0 to 0.9). In each structure pair, one structure is from H. sapiens, and the other is from E. coli. Note that these two figures illustrate the correlation between structural similarity and sequence similarity or identity. The stratified sampling approach used here means that the figures do not represent the actual distribution of the structural similarity and sequence similarity or identity. For the list of selected experimental structures, see Supplemental Table S1. For the actual distribution at a genomic level, see Figure 3, D through F.











