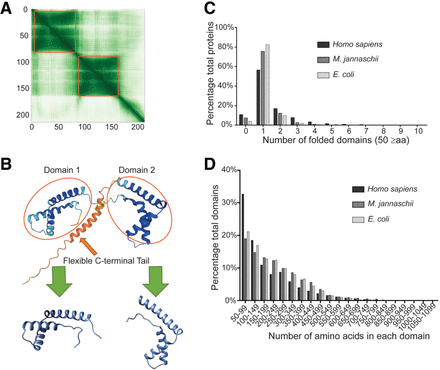
Analysis of the AlphaFold2-predicted structures reveals distinct statistical characteristics between the proteomes of the three model organisms. (A,B) An example illustrating how AlphaFold modeled structures were trimmed into single domains. The orange box indicates the range for each domain. The boundaries of the orange boxes were determined using the Leiden algorithm. (A) The alignment error matrix of the protein HMGB1 (UniProt ID P09429). The axes represent the positions of residues along the sequence of the protein. The x-axis corresponds to the reference position, and the y-axis corresponds to the predicted position. The color at the coordinate (x,y) indicates the AlphaFold2's expected position error of residue y if residue x serves as the reference and the predicted and true positions of residue y are compared. Darker color indicates lower errors or higher accuracy, whereas lighter color indicates higher error or lower accuracy. Thus, the matrix is an indication of the confidence of the relative position between any of the two residues. (B) The modeled tertiary structure of HMGB1 and its relationship with the two individual domains. Blue color indicates high model confidence, and orange color indicates low model confidence. (C) Distribution of the number of domains in each protein structure for all the modeled protein structures from each of the three species in the AlphaFold2 database. Unfolded domains and domains smaller than 50 aa were excluded from the analysis. (D) Distribution of the number of amino acids in each domain for all the modeled protein structures in the AlphaFold2 database. Unfolded domains and domains smaller than 50 aa were excluded from the analysis











