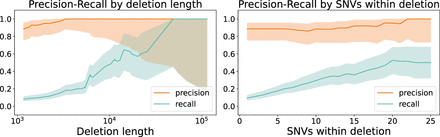
Precision and recall of inherited deletions for HG002. Inherited deletions detected by PhasingFamilies show high precision, with recall dependent on the length of the deletion. As deletion length increases, recall increases from 0.082 to 1.0, as shown in the left panel. Recall also improves with more SNVs within the deleted region, as shown in the right panel. Shaded regions indicate the Agresti–Coull binomial confidence interval.











