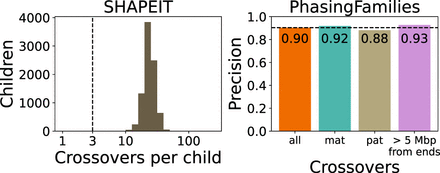
Comparison with SHAPEIT5 on Chromosome 10. The panel on the left shows the number of crossover calls per child made by the haplotype estimation tool SHAPEIT5. The dotted line indicates the number of expected crossovers on Chromosome 10, based on the median value in the platinum genome family NA1281. Although SHAPEIT5 produced far too many false-positive crossover calls to evaluate recall, we were able to validate the precision of PhasingFamilies, as shown in the right panel. Precision was significantly higher for maternal crossovers than for paternal crossovers.











