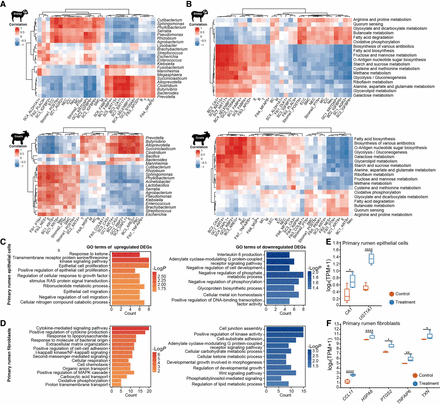
Ruminal microbiota correlations with single-cell transcriptome sequencing in sheep and goats, and transcriptomic analysis of co-cultured rumen cells with Prevotella copri. (A) Correlations of host rumen cell types with the dominant ruminal microbiota. Heatmap shows the Spearman's correlation coefficient between the Eigengene/PC1 value of cell types and the abundances of dominant ruminal microbiota (P < 0.05). (B) Correlations of host rumen cell types with microbial KEGG pathways. Heatmap shows the Spearman's correlation coefficient between the Eigengene/PC1 value of cell types and the abundances of selected microbial KEGG pathways (P < 0.05). The abbreviations of the cell names are shown in Figure 1. (C,D) GO terms (P < 0.05) for up- and down-regulated DEGs (P < 0.05 and |logFC| > 0.75) between the treatment group (co-cultured with P. copri) and control group in primary rumen epithelial cells (C) and primary rumen Fib (D). (E,F) Boxplots showing expression levels of overlap between up-regulated DEGs (treatment vs. control) in primary rumen epithelial cells (E) and primary rumen Fib (F) with up-regulated DEGs (stages P2 vs. P1; Padj < 0.05 and |logFC| > 0.25), which were significantly (P < 0.05) correlated with P. copri. (****) P < 0.0001, (**) P < 0.01, (*) P < 0.05.











