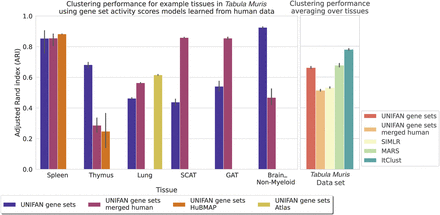
Performance on the Tabula Muris data when using gene set activity scores models pretrained on human tissues. The plot on the left shows the ARI for some example tissues, and the one on the right shows the average ARI across tissues. All versions of UNIFAN methods use models pretrained on human tissues except for “UNIFAN gene sets,” which used models trained on the same data sets as we discussed before. “UNIFAN gene sets merged human” uses the model pretrained on all available human tissues. “UNIFAN gene sets HuBMAP” uses the model pretrained on the corresponding HuBMAP tissue (HuBMAP spleen or thymus). “UNIFAN gene sets Atlas” uses the model pretrained on the “Atlas lung” data set. We included only the best-performing prior methods on the Tabula Muris data (SIMLR, MARS, ItClust) for comparison. We see that the model pretrained on human data is helpful for mouse gene set activity scores inference and for clustering, specifically for tissues having similar cell types between human and mouse such as spleen and lung. For thymus and Brain_Non-Myeloid, whose cell types are not well represented in the pretraining set, the performance drops.











