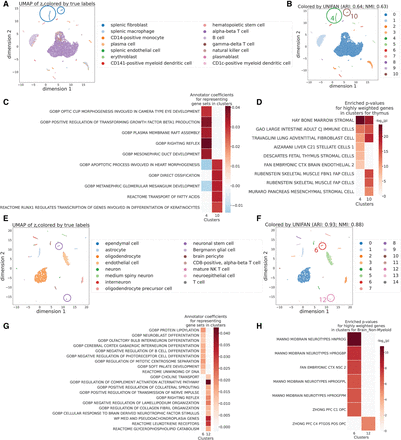
UNIFAN identifies novel cell subtypes for the HuBMAP thymus data and the Brain_Non-Myeloid data in Tabula Muris. (A–D) Results for the HuBMAP thymus data. (E–H) Results for the Brain_Non-Myeloid data in Tabula Muris. (A,B,E,F) UMAP visualization of the low-dimensional representation ze of cells from UNIFAN. Cluster 4 and 10 are circled for the HuBMAP thymus data. Cluster 6 and 12 are circled for the Brain_Non-Myeloid data in Tabula Muris. (A,E) Colored by original cell type labels. (B,F) Colored by the clusters identified by UNIFAN. (C,G) Coefficients learned by the annotator for highly ranked gene sets for the two clusters. (D,H) Enrichment P-values of cell type marker sets in the highly weighted genes learned by the annotator.











