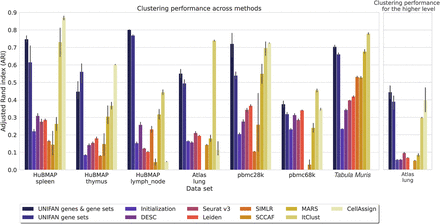
UNIFAN significantly outperforms other methods. “UNIFAN genes & gene sets” is the default UNIFAN version using both gene set activity scores and a subset of genes as features for the annotator. “UNIFAN gene sets” uses only the gene set activity scores. “Initialization” is the initialization clustering results. The others are the prior methods we used for comparison. For the Tabula Muris data, we take the average over all tissues. For tissue-specific results, see Supplemental Figures S11 and S12. The “Atlas lung” data provide two levels of cell type annotations, and so, we show results for both (less detailed annotation comparison shown on the right). SIMLR was unable to cluster the “pbmc68k” and “Atlas lung” data because it ran out of memory. CellAssign does not have an average over all tissues for “Tabula Muris” because some of the tissues in that data set do not have matched cell type marker genes. For details, see Supplemental Methods.











