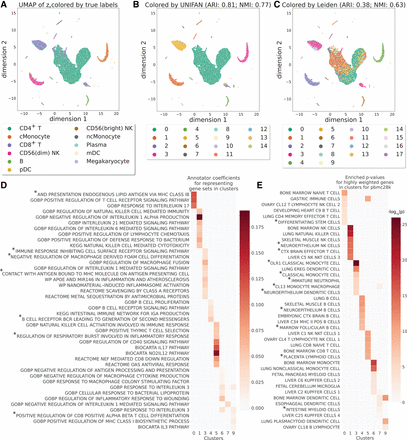
UNIFAN accurately clusters cells and correctly identifies biological processes/pathways. Results presented for the “pbmc28k” data set. (A–C) UMAP visualization of the low-dimensional representation ze of cells output from UNIFAN: (A) colored by true cell type labels, (B) colored by the clusters found by UNIFAN, and (C) colored by Leiden clustering. (D) Coefficients learned by the annotator for highly ranked gene sets for some of the clusters. (E) Enrichment P-values of cell type marker sets in the highly weighted genes learned by the annotator. Here we show the result from the best run for both UNIFAN and Leiden. Because of space limits, some gene set names in D and E are truncated (marked with *). For the full names, see Supplemental Tables S2 and S3.











