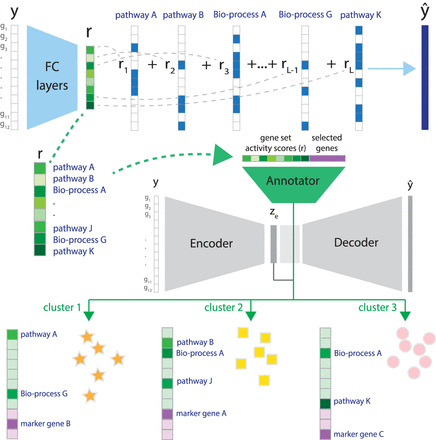
Overview of UNIFAN. (Top) Using the expression levels for genes in a cell y, UNIFAN first infers gene set activity scores (r), using an autoencoder. The decoder is composed of binary vectors with values indicating if a gene belongs to a known gene set or not. (Middle) Next, UNIFAN clusters cells by using the learned gene set activity scores and a low-dimensional representation of the expression of all genes in the cell (ze). For this, it uses an autoencoder-based neural network, which contains two parts: the cluster assignment part (gray) and the “annotator” (green). The cluster assignment part assigns a cell to clusters based on the low-dimensional representation (ze), whereas the “annotator” refines clustering and annotates clusters with biological processes and highly variable genes. (Bottom) Cells assigned to different clusters characterized by selected gene sets and genes. (FC layers) Fully connected layers; (Bio-process) biological process.











