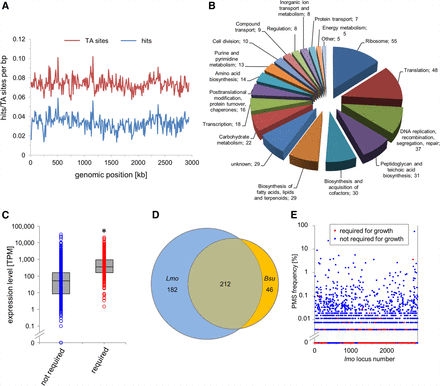
Identification of L. monocytogenes genes required for growth in BHI broth by Tn-Seq. (A) Distribution of TA sites and Tn insertion sites along the L. monocytogenes EGD-e genome. TA sites/hits per base pair were calculated using a coverage window of 10,000 bp. (B) Functional classes of the 394 L. monocytogenes EGD-e genes that are required for growth in BHI broth at 37°C according to Tn-Seq. (C) Expression levels of the 394 genes required for growth in comparison to all other L. monocytogenes EGD-e genes according to a previously published RNA-seq experiment also performed under standard growth conditions (Hauf et al. 2019). The asterisk indicates statistical significance (t-test, P < 0.0001). (D) Venn diagram illustrating the intersection between the 394 L. monocytogenes genes required for growth and the essential genes of B. subtilis 168 (n = 258). (E) Frequency of naturally occurring premature stop codons (PMSs) in each of the L. monocytogenes EGD-e genes as observed in a data set of 27,118 L. monocytogenes genomes from clinical and environmental isolates that were downloaded from the NCBI server.











