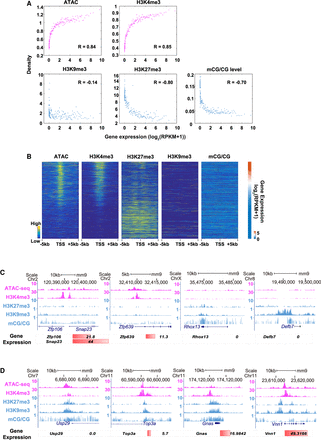
Multifaceted regulation of epigenetic marks at ICM stage. (A) Scatter plots show the correlations between gene expression (GSE66582) and epigenetic signals (ATAC-seq: GSE66581; H3K4me3: GSE71434; H3K9me3: GSE97778; H3K27me3: GSE76687; mCG/CG: GSE56697) in promoters (2 kb upstream of TSSs). Correlation coefficients were calculated by Pearson's linear correlation. (B) Heatmaps show the enrichment of epigenetic marks (ATAC-seq: GSE66581; H3K4me3: GSE71434; H3K9me3: GSE97778; H3K27me3: GSE76687; mCG/CG: GSE56697; normalized RPKM) in the ICM stage; a bar chart shows the expression level of corresponding genes. (C,D) UCSC plots show epigenetic signals (ATAC-seq: GSE66581; H3K4me3: GSE71434; H3K9me3: GSE97778; H3K27me3: GSE76687; mCG/CG: GSE56697) surrounding Zfp106, Snap23, Zfp639, Rhox13, Defb7, Usp29, Top3a, Gnas, and Vnn1. The bar plot shows gene expression level.











