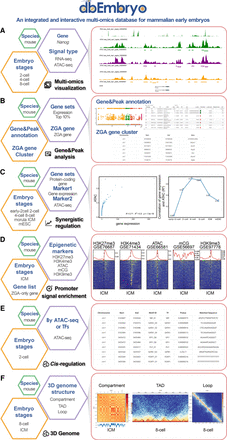
The main functions of dbEmbryo. (A) “Multi-omics visualization” searches and browses the tracks of epigenetic signals by gene names or chromosomes. (B) “Gene&Peak analysis” contains two functions. “Gene&Peak annotation” performs GO and KEGG functional annotation of specific gene sets or peaks for epigenetic signals. “ZGA gene cluster” defines different gene clusters for ZGA genes to represent different synergistic epigenetic mechanisms. (C) “Synergistic regulation” examines the correlation of different epigenetic signals with gene expression at various developmental stages. (D) “Promoter signal enrichment” compares the enrichment of different epigenetic signals at gene promoters spanning developmental stages. (E) “Cis-regulation” identifies transcription factor (TF) binding sites (TFBSs) based on ATAC-seq or TFs. (F) “3D genome” visualizes the 3D genome information (e.g., compartment, TAD, loop) across early embryo development stages in different resolutions.











