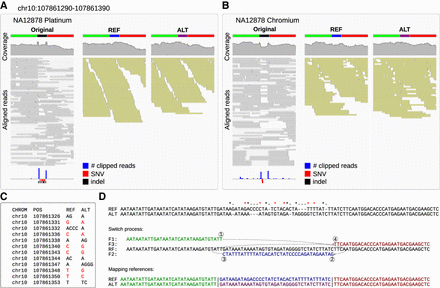
TSM candidates identified with short-read data. (A) In the Platinum data (left), a region in Chromosome 10 has an excess of soft clips (bottom, blue bars) and called variants (SNVs in red, indels in black); the mapping coverage (top, in gray) also shows atypical patterns. (B) Similar signals are seen in Chromium data. (C) The Platinum variant data contain six SNVs and six indels within 28 bp. (D) De novo assembly of the reads creates two locally highly dissimilar haplotypes compatible with the called variants (top). All differences can be explained with a TSM event, an inversion in place (middle). Using two haplotypes with alternative central parts (blue and magenta; bottom) as the reference, extracted reads map in full length (A,B; middle, right) with roughly even coverages. No variants for NA12878 were called in this region in HaplotypeSV data.











