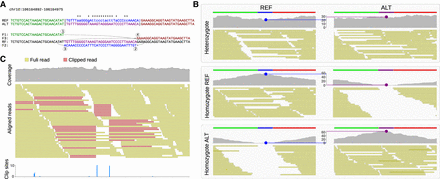
Genotyping of a TSM locus. (A) TSM solution for a complex mutation on Chromosome 10. The REF and ALT alleles share the left (green) and right (red) flanking region but have a different central part (blue and magenta). The differences in the central part are explained by a TSM event (below). (B) The two haplotypes with alternative central parts (blue and magenta bars) are used as the reference for remapping of reads extracted from the locus. In a heterozygous individual (top), reads are mapped evenly to the two alleles, giving uniform coverages (gray). In individuals homozygous for the REF allele (middle) and ALT allele (bottom), reads covering the central part are mapped predominantly to one allele only. Genotype is inferred of the mapping coverage in the middle of the locus (blue and magenta dots and dotted lines) using a function similar to that for genotype calling on nucleotides. (C) NA12878 Platinum data show fairly normal mapping coverage (gray, top), but a closer look at the reads (full and clipped reads in yellow and red, respectively) reveals an anomaly. The cluster of clip sites (blue; bottom) allows computational identification of the locus.











