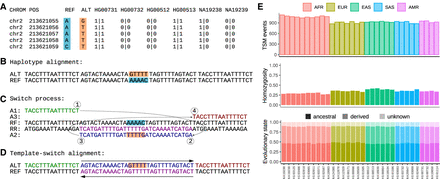
TSMs in HaplotypeSV data. (A) HaplotypeSV data contain a cluster of five SNVs that segregate as a unit among individuals (the first six are shown). (B) Within their genomic context, REF (cyan) and ALT (orange) alleles create two haplotypes. (C) The FPA algorithm attempts to explain the observed differences with two reciprocal template-switch events. In this case, the ALT sequence (A1, A2, A3; shown in green, blue, and red) can be created from the REF sequence by copying the A2 fragment in reverse-complement (RR; shown in magenta). (D) The template-switch solution fully explains the cluster of five base differences. (E) Total numbers of inferred TSM events in different HaplotypeSV samples after removal of low-complexity sequences (top); proportion of homozygous loci (middle); and proportion of ancestral (dark), derived (intermediate), and unpolarized (light) alleles (bottom).











