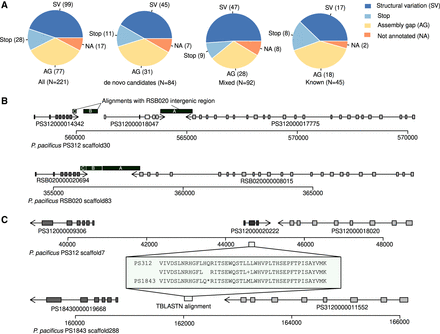
Nonsense mutations and structural variations are the major source of gene loss. (A) The pie charts show the classification of missing orthologs after manual inspection. De novo candidates were defined as genes that are specific to the pacificus clade. (B) The plots show syntenic regions surrounding the Pristionchus-specific gene PS312000018047 in two P. pacificus strains. The x-axis indicates the nucleotide position; exons are shown as rectangles; and syntenic orthologs are color-coded. The presence of two intergenic blocks between the syntenic anchor genes without homology suggests that at least two structural variations occurred that have led to the loss of PS312000018047. (C) The plot shows syntenic regions surrounding the gene PS312000020222 in two P. pacificus strains. The gene is Pristionchus specific and is present in all P. pacificus genomes except in the PS1843 assembly, where it has acquired a premature stop codon.











