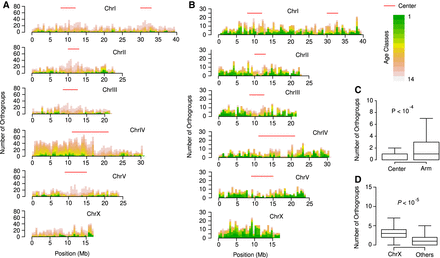
New gene classes are nonuniformly distributed across chromosomes. (A) The number of known orthogroups is plotted in nonoverlapping 500-kb windows across the P. pacificus genome. Center-like regions were defined based on genetic diversity (Rödelsperger et al. 2017) and are indicated by red lines. (B) The plot shows the distribution of de novo candidates across the P. pacificus chromosomes. (C) The box plot shows the number of de novo candidates per 500-kb window. De novo candidates of age class 1 are significantly depleted from chromosome centers. (D) De novo candidates of age class 1 are significantly enriched on P. pacificus Chromosome X.











