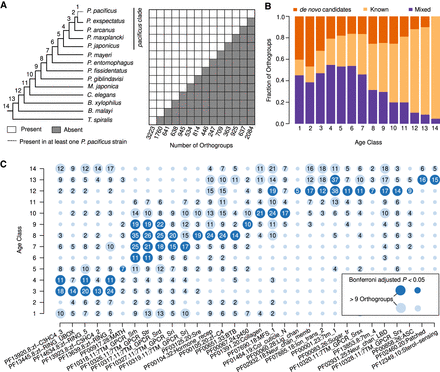
Duplications and new gene formation shape genome evolution. (A) The presence/absence patterns of species in a given orthogroup were used to assign orthogroups to 14 distinct age classes. The age classes define a branch in the schematic phylogeny (Rödelsperger et al. 2018; Smythe et al. 2019), at which a given orthogroup evolved. (B) Orthogroups were classified as known if more than half of the genes had recognizable protein domains and classified as de novo candidates if there was no BLASTP hit against sequences from a set of outgroup species. The remaining orthogroups were classified as mixed. Although older age classes are dominated by members of known gene families, younger age classes are biased toward de novo candidates. (C) The circles represent the number of orthogroups with a given protein domain, and the color code indicates significant enrichments within an age class.











