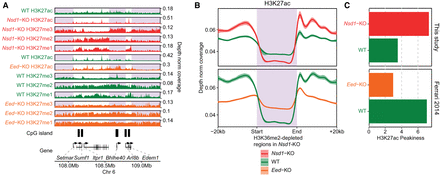
H3K27 methylation controls pervasive H3K27ac. (A) Comparison of H3K27 acetylation in Nsd1-KO versus WT and Eed-KO versus WT, showing the decrease in pervasive acetylation in Nsd1-KO and increase in Eed-KO. The shaded areas indicate H3K36me2-loss (or H3K27me3-spreading) regions in Nsd1-KO. The tracks are only normalized by read depth, not by any quantitative ratios, because no spike in or MS public data are available, and the distributions only reflect changes in levels between relevant regions. (B) Aggregate H3K27ac signal (read-depth normalized, not quantitatively) around H3K36me2-loss regions (n = 4361) in Nsd1-KO, showing the relative reduction in H3K27ac in Nsd1-KO and increase in Eed-KO, in those H3K36me2-loss regions compared to surrounding regions. (C) Peakiness scores of H3K27ac, showing the reduction of background H3K27ac (i.e., more restricted distribution or higher Peakiness score) in Nsd1-KO, and gain of background signal (less localized or lower Peakiness score) in Eed-KO. Comparison of H3K27ac in Ezh2-KO and WT (mESC in 2i medium) is shown in Supplemental Figure S4A–C. The ChIP-seq of H3K27ac and H3K27me in Nsd1-KO and WT are generated in this study, but those in Eed-KO and WT are from Ferrari et al. (2014).











