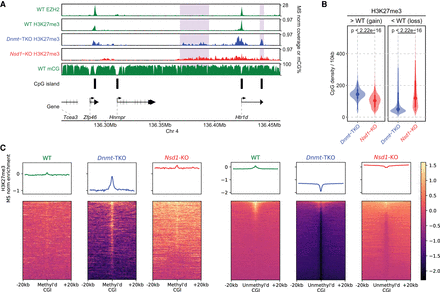
The DNAme-dependent H3K27me3 changes are more CpG-focused than those dependent on H3K36me2. (A) H3K27me3 in WT versus Dnmt-TKO versus Nsd1-KO, showing (in Dnmt-TKO) new peaks around CpG-rich regions and CGIs that were highly methylated and had no PRC2 binding in the WT (shaded areas). (B) Comparison of CpG density in regions gain/loss H3K27me3 in Nsd1-KO and Dnmt-TKO, indicating the H3K27me3 gain in TKO occurs in regions with elevated CpG density. P-values are based on t-test. A similar plot using replicates is provided in Supplemental Figure S3A. (C) Signal intensity plot of H3K27me3 (normalized by input and MS ratios) centered on highly methylated CGIs (n = 516) and unmethylated CGIs (n = 7400) in WT, respectively. In Dnmt-TKO H3K27me3 is gained within CGIs that were highly methylated in WT, but lost from its normal PRC2 nucleation sites, which had low DNAme in WT. Such changes do not occur in Nsd1-KO. Replicate plots are provided in Supplemental Figure S3B, and additional analysis in Supplemental Figure S3C,D. Data used in this figure are generated in this study, except EZH2 ChIP-seq data are from Kundu et al. (2017).











