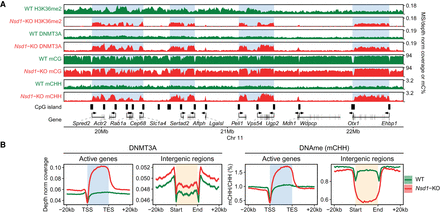
Nsd1-KO reduces DNAme (mCG) and redistributes DNMT3A and de novo DNAme (mCHH). (A) Genome browser representation of H3K36me2, DNMT3A, mCG, and mCHH in WT (green) versus Nsd1-KO (red), showing redistribution of H3K36me2 to genes, followed by DNMT3A and de novo DNAme. The shaded areas indicate H3K36me2-enriched regions in Nsd1-KO. (B) Aggregate signals of DNMT3A and mCHH around active genes (n = 15,850) and intergenic regions (n = 11,629), showing the enrichment (Nsd1-KO compared to WT) in active genes and depletion in intergenic regions. Data used in this figure are generated in this study.











