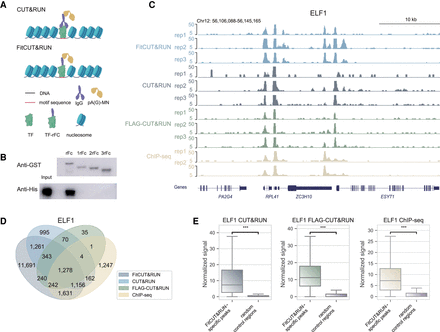
Feasibility and robustness of FitCUT&RUN with K562 cells. (A) Adaptation of FitCUT&RUN based on the common CUT&RUN method. (B) Western blot analysis of the in vitro pull-down assay to determine the interaction of pAG-MN with different truncated rFc fragments. (C) The genome browser view shows ELF1 FitCUT&RUN, CUT&RUN, FLAG-CUT&RUN (three replicates for each), and ChIP-seq (two replicates) signals around RPL41 loci as a representative example of the consistency among these methods. (D) Venn diagram showing the overlap status between ELF1 FitCUT&RUN, CUT&RUN, FLAG-CUT&RUN, and ChIP-seq peaks. The majority of ChIP-seq, CUT&RUN, and FLAG-CUT&RUN peaks were also detected by FitCUT&RUN. (E) Boxsplots showing that the normalized ELF1 ChIP-seq, CUT&RUN, and FLAG-CUT&RUN signal on FitCUT&RUN-specific peaks is significantly greater than that on random control regions, confirming the reliability of the FitCUT&RUN data.











