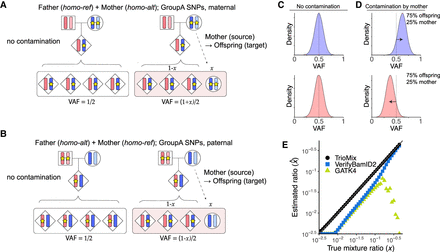
Quantification of offspring DNA contamination by parents. GroupA single nucleotide polymorphisms (SNPs) are used for quantifying parental DNA contamination in the DNA mixture. SNPs are shown as stars; each bar represents a chromosome in the diploid genomes. (A) SNP loci in which the mother has the homo-alt genotype are shown (maternally inherited GroupA SNPs). When the offspring's DNA is not contaminated, the variant allele frequency (VAF) of the offspring is 50%. When the mother's contamination level is x, the expected offspring VAF in the contaminated sample is (1+x)/2. (B) A similar analysis can be performed with SNP loci in which the father has the homo-alt genotype (paternally inherited GroupA SNPs). (C) A density plot of VAFs for maternally (blue) and paternally (red) inherited GroupA SNPs is shown when there is no contamination (offspring 100%). (D) A density plot of VAFs for maternally (blue) and paternally (red) inherited GroupA SNPs with 25% maternal contamination is shown. (E) True versus estimated value for TrioMix (black circle), VerifyBamID2 (blue square), and GATK4 CalculateContamination (green triangle) for in silico simulated DNA contamination with parental DNA. Estimated values below 10−2.5 are shown on the plot's x-axis.











