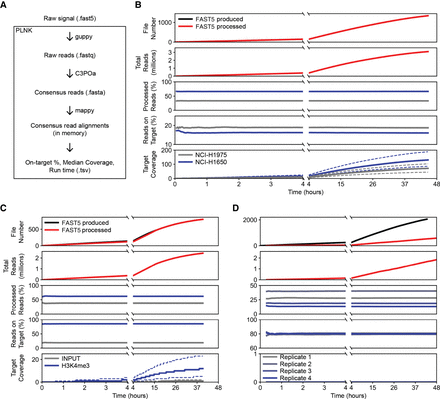
Real-time characterization of Illumina sequencing libraries. (A) Diagram of PLNK functionality; FAST5 files processed in the order they are produced. PLNK controls guppy5 for base-calling, C3POa for consensus calling, and mappy for alignment, as well as calculates metrics based on those alignments. (B–D) Simulation of real-time analysis for enriched Tn5 (B), ChIP-seq (C), and RNA-seq (D) libraries. For each time point, panels from top to bottom show (1) the number of FAST5 files that are produced and processed, (2) the number of demultiplexed reads produced by guppy5/C3POa/demultiplexing, (3) the percentage of reads associated with each library in the sequenced pool, (4) the percentage of reads overlapping with target regions, and (5) the median read coverage of bases in the target regions.











