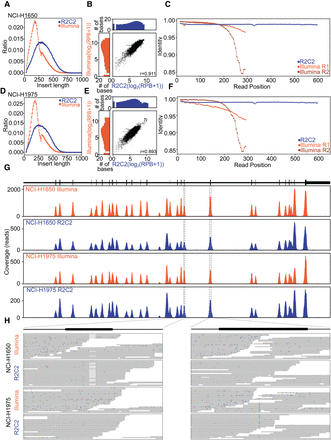
Evaluating target-enriched Tn5 libraries with R2C2. (A,D) Insert length of library molecules sequenced by Illumina or R2C2 approaches. (B,E) Comparison of per-base coverage in the Illumina and R2C2 data sets. Marginal distributions are log2 normalized. (C,F) Alignment-based read position–dependent accuracy shown for the indicated sequencing reads and methods. (G,H) Sequencing coverage plot of the target-enriched Tn5 libraries for R2C2 and Illumina results at Chromosome 7: 55,134,584–55,211,629, which covers a part of the EGFR gene. Top panel shows the annotation of one EGFR isoform. The x-axis of the coverage plot is the base pair position, and the y-axis is the total number of reads at each position. The dotted lines indicate zoomed-in views of exons that contain the 15-bp deletion in NCI-H1650 (left) and the C-to-T and T-to-G point mutations in NCI-H1975 (right). Both samples’ Illumina reads and the R2C2 read alignments of the selected regions are shown. The mismatches are colored based on the read base (A, orange; T, green; C, blue; G, purple).











