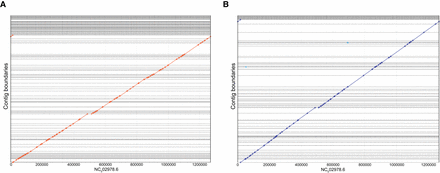
Comparing R2C2 and Illumina based assemblies of a small genome. Illumina 2 × 150 reads were assembled in 134 contigs using Meraculous. R2C2 reads were assembled using miniasm into 95 contigs. The alignments of the contigs of both assemblies—(A) Illumina and (B) R2C2—are shown as dot plots generated by MUMmer (Kurtz et al. 2004). Both approaches failed to assemble a section of the Wolbachia genome that contains pseudogenes and a transposable element near coordinate 500,000.











