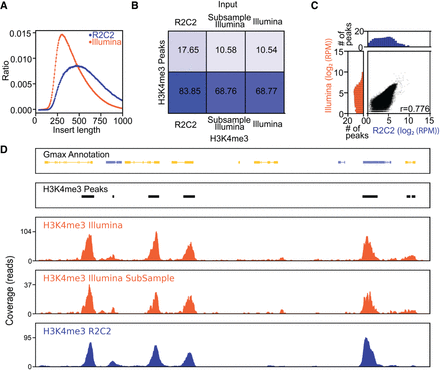
Sequencing ChIP-seq libraries on the ONT MinION after R2C2 conversion. (A) Insert length distribution of R2C2 and Illumina NovaSeq 6000 reads generated from the same Illumina library. (B) Percentage of reads in the R2C2, subsampled Illumina, and full Illumina data sets overlapping with H3K4me3 peaks generated from the full Illumina H3K4me3 data set using MACS2. (C) The comparison of the number of R2C2 and subsampled Illumina reads overlapping with H3K4me3 peaks is shown as scatter plots with marginal distributions shown as histograms. Pearson's r is shown at the bottom right. (D) Genome annotation, H3K4me3 peak areas, and read coverage histograms are shown for a section of the Gmax genome.











