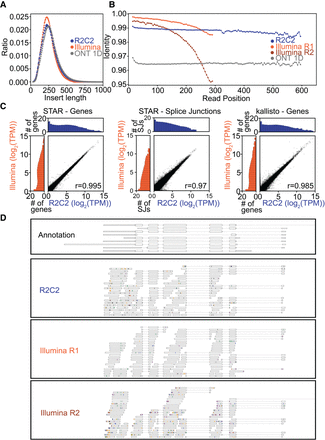
Sequencing Illumina RNA-seq libraries on the ONT MinION after R2C2 conversion. Insert length distribution (A) and read position–dependent identity to the reference genome (B) of R2C2 and Illumina MiSeq reads generated from the same Illumina library. (C) Comparisons of R2C2 and Illumina MiSeq read-based gene expression and splice junction usage quantification by STAR and kallisto are shown as scatter plots with marginal distributions (log2 normalized) shown as histograms. (D) Genome browser-style visualization of read alignments to the Actb locus. Mismatches are marked by lines colored by the read base (A, orange; T, green; C, blue; G, purple). Insertions are shown as gaps in the alignments, and deletions are shown as black lines.











