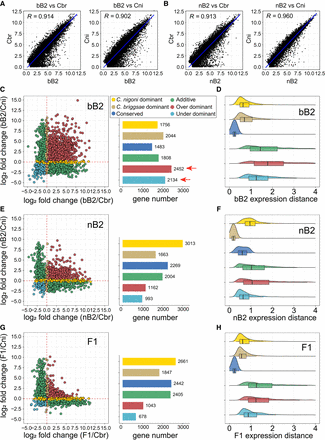
bB2 embryos show a higher degree of transgressivity in expression inheritance profile of coding genes relative to other types of hybrid embryos. (A,B) Pairwise comparisons of the normalized expression levels (read counts) of the one-to-one orthologs between bB2 (A) or nB2 (B) and parental embryos. Also shown is Pearson's correlation coefficient (R). Linear regression lines are shown in blue. (C,E,G) Scatter (left) and bar plots (right) showing the expression level change of the orthologs in the bB2 (C), nB2 (E), or F1 (G) embryos compared with that of each parental species, that is, Caenorhabditis briggsae (x-axis) and Caenorhabditis nigoni (y-axis). Inheritance categories are differentially color coded as indicated. Red dashed lines indicate the “0” of log2 fold change for each axis. The total gene number of each inheritance category in the three hybrids is shown. The numbers of over- and under-dominant are highlighted with arrows. (D,F,H) Density plots with overlaid box plots showing the expression distance (see Methods) of one-to-one orthologs within each inheritance category for bB2 (D), nB2 (F), and F1 (H) embryos.











