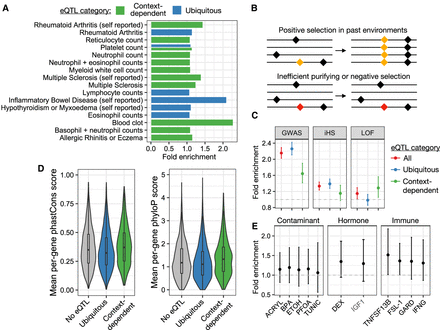
Phenotypic relevance and evolution of context-dependent eQTLs. (A) Diseases and heath conditions for which associated genes (identified via PTWAS) (Zhang et al. 2020) are enriched among genes with one or more context-dependent or ubiquitous eQTLs. The x-axis represents the fold enrichment estimate from a Fisher's exact test. (B) Evolutionary forces that potentially maintain context-dependent eQTLs: (1) positive selection on beneficial mutations (yellow) or (2) inefficient purifying or negative selection that fails to remove deleterious mutations (red). (C) Overlap of ubiquitous, context-dependent, and all (ubiquitous and context-dependent) eQTLs with (1) the full catalog of GWAS-associated loci (Hindorff et al. 2009), (2) iHS outlier loci (Johnson and Voight 2018), and (3) genes annotated as loss of function and mutation intolerant (Lek et al. 2016). The y-axis represents fold enrichment from a Fisher's exact test, with error bars denoting the 95% confidence interval for each estimate. (D) Distribution of mean per-gene phastCons and phyloP scores for genes with no eQTLs, one or more ubiquitous eQTLs, and one or more context-dependent eQTLs. (E) Overlap of response eQTLs, identified in a given condition, with iHS outlier loci (Johnson and Voight 2018). The y-axis represents fold enrichment from a Fisher's exact test, with error bars denoting the 95% confidence interval for each estimate.











