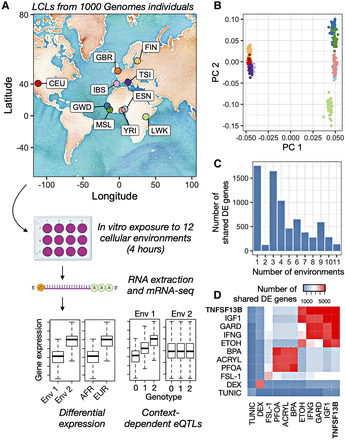
Study overview and environmental effects on gene expression. (A) Lymphoblastoid cell lines (LCLs) derived from individuals included in the 1000 Genomes Project were obtained from Coriell Institute. Specifically, we obtained lines derived from individuals of European and African ancestry as noted on the map (abbreviations for included populations are as follows: [CEU] Utah residents [CEPH], [GWD] Gambian Mandinka, [GBR] British, [IBS] Iberian, [MSL] Mende, [FIN] Finnish, [TSI] Toscani, [ESN] Esan, [YRI] Yoruba, and [LWK] Luhya) (see also Supplemental Table S1). Each cell line was exposed to 12 cellular environments for 4 h, after which we harvested the RNA and performed mRNA-seq. These data were used to understand differential expression as a function of environmental context and ancestry ([AFR] African, [EUR] European), as well as to map ubiquitous and context-dependent eQTLs. (B) Principal components analysis of genotype data for individuals included in this study (colors are as in A). Individual used in this analysis are those for which paired RNA-seq and genotype data were available (Supplemental Table S4). (C) Number of differentially expressed (DE) genes shared between N environments using a mashR, joint analysis approach. N is plotted on the x-axis and ranges from one (i.e., the gene is DE in response to only one environmental treatment) to 11 (i.e., the gene is DE in response to all 11 environmental treatments) (see also Supplemental Table S5). The low number of genes when N = 2 is driven by a large number of dexamethasone (DEX)-specific genes, such that 93.7% of the N = 1 genes are DEX-specific; when DEX is excluded from the data set, most genes are shared between three or more environments (Supplemental Fig. S8). (D) Number of DE genes shared between a given pair of environmental treatments using a mashR, joint analysis approach. The diagonal represents the number of DE genes in response to the focal environmental treatment. Abbreviations are as in Table 1.











