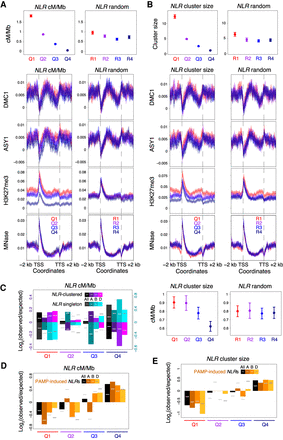
Chromatin, recombination, and clustering at nucleotide-binding and leucine-rich repeat (NLR) genes. (A) NLR genes were divided into four groups corresponding to those in the 100th–75th (Quantile 1), 75th–50th (Quantile 2), 50th–25th (Quantile 3), and 25th–0th (Quantile 4) percentiles with regard to their mean crossover rate (cM/Mb, derived from the Chinese Spring × Renan genetic map) between 1 kb upstream of transcriptional start sites and 1 kb downstream from transcriptional termination sites (Q1–Q4, left), or into four randomized groups (R1–R4, right). Solid circles denote the mean crossover rate for each group of genes (error bars = 95% confidence intervals for mean cM/Mb values). Metaprofiles show windowed mean values (solid lines) for each group of genes and 2-kb flanking regions (transparent ribbons = 95% confidence intervals). ChIP-seq coverage metaprofiles of DMC1, ASY1, and H3K27me3 (IWGSC 2018) are derived from log2(ChIP/input) profiles. (B) As in A, but with NLR genes divided into four groups according to decreasing physical cluster size (Q1–Q4, left), or into four randomized groups (R1–R4, right). Solid circles denote the mean cluster size (upper plots) or crossover rate (lower plots) for each group of genes (error bars = 95% confidence intervals for mean cluster size or cM/Mb values). (C) NLR gene quantiles defined by decreasing mean crossover rate (Q1–Q4 = high–low cM/Mb) were evaluated for representation of physically clustered or singleton NLR genes. Log2(observed/expected) ratios (bar graphs) and significance thresholds (horizontal gray lines, α = 0.05) were calculated by sampling from the hypergeometric distribution 100,000 times. Analyses were performed across all subgenomes and within each subgenome (All, A, B, and D). (D) As in C, but analyzing NLR gene quantiles defined by decreasing mean crossover rate for representation of NLR genes that were up-regulated upon challenge with pathogen-associated molecular patterns (PAMPs) (Steuernagel et al. 2020). (E) As in D, but analyzing NLR gene quantiles defined by decreasing physical cluster size.











