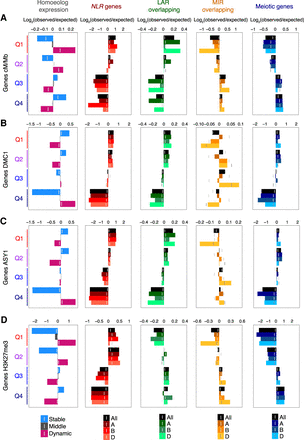
Gene expression pattern, function, and signatures of adaptation according to crossover rate, DMC1, ASY1, or H3K27me3. (A) Gene quantiles defined by decreasing mean crossover rate (Q1–Q4 = high–low cM/Mb) were evaluated for representation of (1) genes assigned to homoeolog expression categories (Homoeolog expression) (Ramírez-González et al. 2018), (2) genes encoding nucleotide-binding and leucine-rich repeat proteins (NLR genes) (Steuernagel et al. 2020), (3) genes overlapping genomic regions associated with local adaptation (LAR overlapping) (He et al. 2019), (4) genes overlapping genomic regions associated with modern wheat improvement (MIR overlapping) (He et al. 2019), and (5) genes with putative functions in meiosis (Meiotic genes) (Alabdullah et al. 2019). Log2(observed/expected) ratios (bar graphs) and significance thresholds (vertical gray lines, α = 0.05) were calculated by sampling from the hypergeometric distribution 100,000 times. Analyses were performed across all subgenomes and within each subgenome (All, A, B, and D). (B) As in A, but with genes divided into four quantiles according to decreasing mean DMC1 log2(ChIP/input) coverage. (C) As in A, but with genes divided into four quantiles according to decreasing mean ASY1 log2(ChIP/input) coverage. (D) As in A, but with genes divided into four quantiles according to decreasing mean H3K27me3 log2(ChIP/input) coverage (IWGSC 2018).











