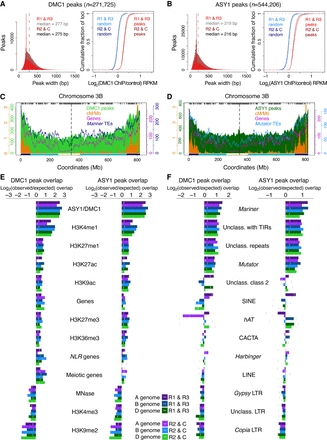
DMC1 and ASY1 ChIP-seq peaks associate with markers of euchromatin and heterochromatin. (A) Histograms of DMC1 ChIP-seq peak widths (bp) in distal compartments (R1 and R3, red) and in interstitial and proximal compartments (R2 and C, burgundy), with median widths indicated by vertical dashed lines in gray and black, respectively. Also shown are cumulative distribution curves for compartmentalized DMC1 ChIP-seq peaks and randomly positioned loci ranked by log2(ChIP/input) RPKM values. (B) As in A, but for ASY1 ChIP-seq peaks and randomly positioned loci. (C) Chromosome 3B profiles of DMC1 ChIP-seq peak frequencies (light green shading), crossover rate (cM/Mb, orange shading) (IWGSC 2018), gene frequency (magenta line), and Mariner transposon frequency (navy line) in 10-Mb sliding windows with a 1-Mb step. Previously defined coordinates delimiting distal (R1 and R3, navy boxes), interstitial (R2a and R2b, turquoise boxes), proximal (C, cream boxes), and centromeric (vertical dashed lines) regions are indicated along the x-axis (IWGSC 2018). Gray ticks denote genetic markers used to construct the Chinese Spring × Renan genetic map. (D) As in C, but showing ASY1 ChIP-seq peak frequency (dark green shading) and Mutator transposon frequency (light blue line). (E) Log2(observed/expected) overlap of compartmentalized DMC1 and ASY1 ChIP-seq peaks in each subgenome with the indicated genomic features, based on the numbers of feature-overlapping loci across 10,000 sets of randomly positioned loci. Vertical gray lines mark significance thresholds (α = 0.05). (F) As in E, but showing log2(observed/expected) overlap of DMC1 and ASY1 ChIP-seq peaks with transposon superfamilies.











