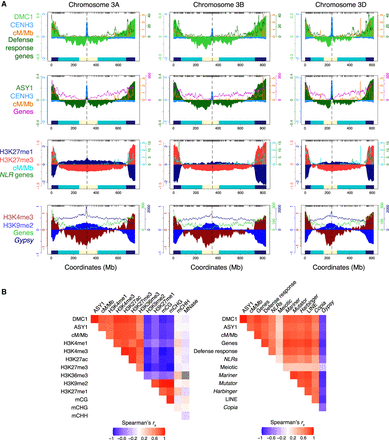
Genomic landscapes of DMC1 recombinase, the meiotic axis protein ASY1, crossovers, and chromatin states in wheat. (A) Coverage profiles along a representative set of homoeologous chromosomes (3A, 3B, and 3D) for DMC1 (light green), ASY1 (dark green), CENH3 (light blue) (Guo et al. 2016), H3K27me3 (red) (IWGSC 2018) (all log2[ChIP/input], using a Chinese Spring chromatin input sequencing library with accession SRR6350669) (IWGSC 2018), H3K27me1 (navy), H3K4me3 (dark red), and H3K9me2 (blue) (all log2[ChIP/MNase], using a Chinese Spring MNase-seq library from this study) ChIP-seq in 1-Mb adjacent windows, smoothed by applying a moving average. ChIP-seq chromosome profiles (shading) are overlaid with crossover rates (cM/Mb, derived from a Chinese Spring × Renan genetic map) (IWGSC 2018), and gene and Gypsy LTR retrotransposon frequencies in 10-Mb sliding windows with a 1-Mb step (lines). A representative set of the IWGSC RefSeq v1.1 Chinese Spring annotation of high-confidence protein-coding gene models (IWGSC 2018), genes annotated with the “defense response” Gene Ontology (GO) term (Ramírez-González et al. 2018), genes encoding nucleotide-binding and leucine-rich repeat (NLR) proteins (Steuernagel et al. 2020), and Gypsy LTR retrotransposons in the IWGSC annotation (IWGSC 2018) were plotted. Previously defined coordinates delimiting distal (R1 and R3, navy boxes), interstitial (R2a and R2b, turquoise boxes), proximal (C, cream boxes), and centromeric (vertical dashed lines) regions are indicated along the x-axis (IWGSC 2018). Gray ticks denote genomic coordinates of genetic markers used to construct the Chinese Spring × Renan genetic map. (B) Genome-wide Spearman's rank-order correlation coefficients (rs) for the indicated parameter pairs profiled in 1-Mb adjacent windows, with cells coded according to the color-gradient correlation scale. Gray boxes denote correlation coefficients that are not significant after P-value standardization.











