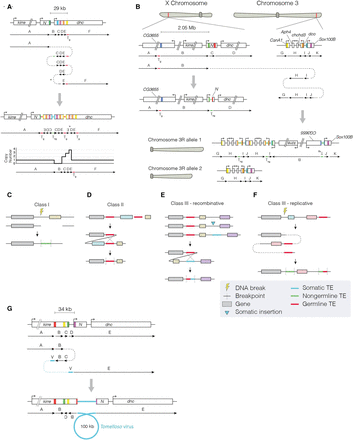
(See figure on preceding page.) Transposable element sequences and viral insertions in Notch-inactivating structural variants. (A,B) Two complex genomic rearrangements inactivating Notch based on read support and CNV calls. Schematics show genomic regions before (top) and after (bottom) each rearrangement. Colored boxes represent breakpoints, with the resulting genomic adjacencies shown below. Arrows indicate the order and orientation of genomic regions. Transposable elements are shown as genomic regions, with nongermline sequences (Tng) shown in green and germline sequences (Tg) shown in red. (A) In sample P63, a complex event generated a deletion in region B, followed by an inverted quadruplication of downstream sequence (regions C, D, and E), flanked by TE sequences. We detected a 12-bp locally templated insertion (indicated by an asterisk) at the breakpoint junction between regions C and D. A schematic of the resulting copy number profile is shown below. (B) In sample P35, a translocation from Notch to Chromosome 3R occurred. A 2.05-Mb region upstream of Notch (region B) was incorporated onto Chromosome 3R, and the entire region was duplicated. The region immediately upstream of the translocation breakpoint on the X Chromosome (region C) was deleted, and we detected TE sequence at the breakpoint, as well as at the 5′ breakpoint of region J, and the junction between regions H and I. In this model, one copy of Chromosome 3R contains the rearranged region from the X Chromosome (labeled allele 1), whereas the other is unaltered (allele 2). We note that other potential configurations may exist for such rearrangements. (C–F) Schematics show putative mechanisms of rearrangement that could explain the signatures of TE involvement detected in Notch-inactivating structural variants. In each class, the uppermost schematic shows a hypothetical genomic region, with genes indicated by colored boxes to help visualize the resulting rearrangement (shown at the bottom). (C) In class I events, read evidence supported a TE or TE fragment integrated at the breakpoint junction. We hypothesize that the TE sequence was integrated during DNA repair. (D) In class II events, two germline TE sequences were found at breakpoint junctions. We hypothesize that these sequences underwent nonallelic homologous recombination, deleting the central region. (E,F) Class III events had evidence for a nongermline TE sequence at one breakpoint junction and germline TE sequence at another. Two interpretations for the breakpoint signatures present in class III events. (E) In the first, a recombinative explanation posits that a de novo TE sequence was inserted (blue arrow) downstream from a germline TE belonging to the same family. Recombination between the two TEs deleted the central region. (F) A second possible explanation of class III breakpoint signatures, wherein DNA damage is repaired by a replicative polymerase that erroneously copies TE sequence into one or several of the breakpoint junctions. This results in a de novo TE signature. (G) A schematic showing genomic regions before (top) and after (bottom) the integration of a fragment of a viral genome in the context of a complex rearrangement detected in sample P31. Although our sequencing data support this configuration, it is possible that alternative explanations exist.











